At least one-third of the stools are bacteria. Are they related to cancer? April 03, 2019 Source: Academic Jingwei Under normal circumstances, one-third to two-thirds of the solid components in human feces are intestinal bacteria. In recent years, there has been increasing evidence that the gut microbiome has an important impact on human health, and more and more scientific studies have attempted to analyze the characteristics of gut microbiota from feces and to learn clues about the disease. In the latest paper published online this week in Nature Medicine, two teams of multinational scientists have made new breakthroughs in this field. They published two papers back-to-back, identifying the gut microbiota characteristics that can be used to predict colorectal cancer from nearly a thousand human fecal samples. They found that although the data used in the two studies were from countries in Europe, Asia, and the Americas, and the stool samples were from people living in different environments and with different eating habits, the gut microbiome had the same color in colorectal cancer. Specific change! One of the tasks was led by scientists at the European Molecular Biology Laboratory (EMBL). Based on the four existing colorectal cancer studies and one new study, they performed a metagenomic analysis of stool samples from 386 patients with colorectal cancer and 392 controls for intestinal health, ie, stool samples. The genetic material of the entire microbial population was analyzed to find out which of the patient's gut flora were significantly different from healthy people. In another work led by Professor Nicola Segata of Trento University in Italy, scientists obtained 969 stool samples from 5 open databases and 2 new cohorts of colorectal cancer patients and healthy controls, using shotgun methods. Metagenome sequencing analysis to find the relationship between gut microbiota and colorectal cancer. Based on the number of fecal samples and the diversity of the population, the researchers believe that these two studies may be the largest colorectal cancer study to date. Since the data comes from many different studies in the past, the sample acquisition methods, storage conditions, DNA extraction methods, etc. are different, and the two groups of scientists have designed optimized statistical methods to eliminate interference factors. In addition, bioinformatics experts use the Metagenome Operational Classification Units (mOTUs) to correlate gene fragments with the microorganisms to which they belong, eliminating the need to isolate and culture bacteria in stool samples, and to identify and quantify bacterial species in the sample. A comprehensive understanding of the gut microbiota. Despite the specific methods used, the two research teams have identified the same bacterial pathogens and strains in the intestinal flora of patients with colorectal cancer that are significantly higher than the healthy intestinal tract, which can be used as a colorectal cancer. Biomarkers. Trento University's research team selected 16 of the most significant bacteria, Dr. Thomas, the lead author of the study, said: "I hope to develop a simple diagnostic tool based on this, without having to sequence the entire microbiota, while at the same time Have the necessary precision." Two findings provide new insights into the development of colorectal cancer. They found that in the intestines of patients with colorectal cancer, in addition to several bacteria known to be associated with intestinal diseases, there is also a very rich bacterium called Fusobacterium nucleatum. Usually live in certain areas of the mouth. In the past, people thought that the strong acidity of gastric juice would prevent these bacteria in the mouth from migrating to the intestines, but it seems to be not the case. This result also suggests that controlling the spread of oral bacteria may also be an important way to regulate the intestinal flora. In addition to identifying significant differences between the bacterial species colonized in the intestines of colorectal cancer patients and healthy intestinal tracts, the research team used a number of databases to train predictive models using machine learning techniques to identify predictive colorectals. Microbial metabolic characteristics of cancer. A pooled analysis of the original metagenomics by the researchers revealed that colorectal cancer is associated with a microbial-degrading choline-related gene. In stool samples from patients with colorectal cancer, the researchers found that the microbial cholestyrase gene (CC) is abundantly present, and the enzyme expressed by this gene is involved in the metabolism of choline. Choline is a nutrient contained in foods such as meat. Certain bacterial species in the intestine convert choline into a metabolite that may be harmful to the human body. This finding further suggests that there may be a carcinogenic link between the gut microbiota and the high-fat diet. The latest nationwide cancer survey shows that colorectal cancer is the third most common malignant tumor in China and the fifth most common malignant tumor. Currently, colonoscopy screening is the most reliable way to control colorectal risk. On the basis of these two studies, it is possible to predict the occurrence of colorectal cancer by examining the microbiological characteristics of the feces in the stool samples in the future, which can reduce the pain of screening and the chance of early detection and early treatment. NINGBO VOICE BIOCHEMIC CO. LTD , https://www.medicine-voice.com

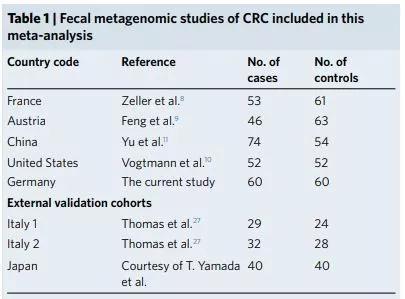
â–²A meta-analysis of one of the studies covered samples from China, the United States, and Europe (Source: References [2]) 
â–² Professor Nicola Segata (10th from left) and his research team at the University of Trento, Italy (Source: University of Trento, filmmaker: Alessio Coser) 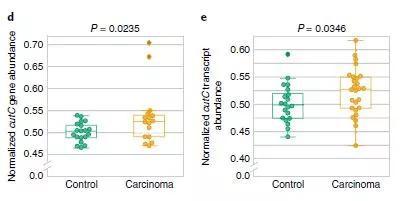
â–²The gene abundance of enzymes related to choline metabolism expressed by intestinal microbes is higher in patients with colorectal cancer (Source: Reference [1])