A. Overview B. tRNA profile and cell fate C. tRNA spectrum and disease Cancer Gingold et al. found that tRNA expression profiles are significantly different between tumor cells and differentiated cells [4]. The expression of tRNA highly expressed in differentiated cells or cells in a resting state is inhibited in proliferating cells. Conversely, tRNAs that are highly expressed in proliferating cells have a lower expression level in differentiated or resting cells. The researchers also found that cancer cells can adjust the tRNA spectrum to facilitate their own development. By comparing the expression levels of tRNA in breast cancer and normal breast tissue, Pavon-Eternod found significant changes in tRNA in the nucleus and mitochondria of breast cancer cells, suggesting that tRNA can be used as a molecular marker for breast cancer [12]. Recently, Goodarzi et al. also demonstrated that certain specific tRNA expression levels are elevated in breast cancer cells during their ability to metastasize. They believe that changes in tRNAGlu-UUC and tRNAArg-CCG expression levels may be one of the predisposing factors for breast cancer metastasis. Further studies indicate that tRNAGlu-UUC can promote the expression of EXOSC2 and GRIPAP1 proteins, thereby promoting tumor metastasis [8]. In conclusion, the disorder of expression of tRNA has a profound effect on the occurrence and development of tumors [5, 8-15]. Gelatin Sheet,Gelatin Sheet Leaf,Halal Gelatin Sheet,Edible Gelatin Sheet Hebei Haodong Biological Technology Co.,Ltd. , https://www.hdgelatin.com
Transfer Ribonucleic Acid (tRNA) is the most abundant short-chain non-coding RNA molecule in the organism. It carries and transports amino acids and participates in protein translation. It is an important bridge connecting mRNA and protein. Although tRNA is widely present in living organisms, different organism genomes have different preferences for specific codons, resulting in differences in tRNA profiles. Codon preference affects translation efficiency and accuracy [1-3]. Therefore, changes in the tRNA profile can have a significant impact on a variety of cellular physiological processes. Arraystar's nrStarTM Human tRNA Repertoire PCR Array contains 66 pairs of nuclear tRNA primers and 22 pairs of mitochondrial tRNA primers, allowing researchers to easily and quickly analyze tRNA profiles. This product contains all the anti-codons provided by GtRNAdb and tRNAdb and is a powerful tool for studying human tRNA profiles.
Studies have shown that changes in the tRNA profile can influence fate decisions during cell development. A series of life activities such as cell proliferation [4], cell differentiation [4, 5] and apoptosis [6] are accompanied by changes in tRNA levels. There are significant differences in codon usage between genes involved in cell maintenance-related genes and cell polymorphisms. The tRNA profile changes induced by cell proliferation and cell differentiation correspond to cell maintenance-related genes and cell polymorphism genes, respectively (Fig. 1). In addition, cell maintenance-related gene mRNA specifically induces expression in proliferating cells and tumor cells, suggesting a direct correlation between transcription and translation [4]. Overexpression of the initiating tRNA (tRNAiMet) can significantly affect the entire tRNA expression profile and further lead to an increase in cellular metabolic capacity and proliferative capacity [5]. tRNA can also affect cytochrome C-mediated apoptotic body formation and participate in the regulation of apoptosis [6]. By microinjection of tRNA in vitro, the researchers found that tRNA can inhibit the cytochrome C-mediated apoptotic pathway [7]. In summary, the tRNA profile can regulate the physiological state of a cell in a variety of ways. 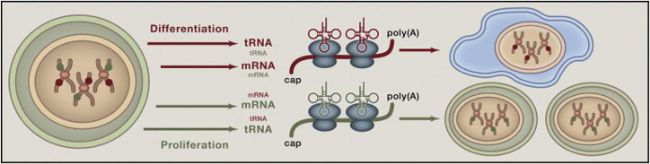
Figure 1. The tRNA profile is adapted to changes in the transcriptome.
The tRNA spectrum not only affects the physiological state of a variety of cells, but also plays an important role in the development of various diseases. For example, the expression disorder of tRNA can induce tumorigenesis [5]. Therefore, tRNA expression profiling research is of great significance for revealing the pathological mechanism of human diseases.
Huntington's disease Huntington's disease is a highly prevalent hereditary neurodegenerative disease caused by an increase in the end of the huntingtin protein polyglutamine. Analysis of the brain tissue of Huntington's disease patients revealed the presence of polyalanine and polyserine traces in polyglutamine. These polyamino acids may be due to a frameshift of the glutamine-encoding CAG reading frame, resulting in a GCA reading frame encoding alanine or an AGC reading frame encoding serine. Girstmair et al. found that insufficient expression of tRNAGln-CUG is the key to inducing transcoding errors. In addition, the ratio of glutamine to alanine affects the morphology of the frameshift translation protein. These results indicate that the expression disorder of tRNA and its resulting frameshift translation may be related to the occurrence of Huntington's disease and the heterogeneity of disease individuals [16].
Viral infections are well known and viral protein synthesis is completely dependent on the host's translation system, including the use of tRNA libraries. It is therefore generally accepted that the virus selectively adapts to the tRNA pool of the host cell. Since the use of host codons determines the host's tRNA library, viral protein synthesis has high translational efficiency only when using codons that are highly similar to the host gene. However, in many cases, the codon usage of the viral gene does not match the host gene. Pavon-Eternod found that influenza A (influenza A) and vaccinia virus can modulate the tRNA pool of infected hosts to adapt to the translation of their own genes [17]. Studies have also shown that the early genes of HIV (HIV-1) are highly similar to the codon usage of host genes, while the late genes of HIV are not. In the late stage of viral infection, the host tRNA library is more suitable for protein translation of HIV [18]. Therefore, the virus can adapt the host tRNA library to adapt to the needs of its own protein synthesis.
references
[1] Drummond DA, Wilke CO. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell 2008;134:341-52.
[2] Plotkin JB, Kudla G. Synonymous but not the same: the causes and consequences of codon bias. Nature reviews Genetics 2011;12:32-42.
[3] Shah P, Gilchrist MA. Explaining complex codon usage patterns with selection for translational efficiency, mutation bias, and genetic drift. Proceedings of the National Academy of Sciences of the United States of America 2011;108:10231-6.
[4] Gingold H, Tehler D, Christoffersen NR, Nielsen MM, Asmar F, Kooistra SM, et al. A dual program for translation regulation in cellular proliferation and differentiation. Cell 2014;158:1281-92.
[5] Pavon-Eternod M, Gomes S, Rosner MR, Pan T. Overexpression of initiator methionine tRNA leads to global reprogramming of tRNA expression and increased proliferation in human epithelial cells. Rna 2013;19:461-6.
[6] Mei Y, Stonestrom A, Hou YM, Yang X. Apoptotic regulation and tRNA. Protein & cell 2010;1:795-801.
[7] Mei Y, Yong J, Liu H, Shi Y, Meinkoth J, Dreyfuss G, et al. tRNA binds to cytochrome c and inhibits caspase activation. Molecular cell 2010;37:668-78.
[8] Goodarzi H, Nguyen HC, Zhang S, Dill BD, Molina H, Tavazoie SF. Modulated Expression of Specific tRNAs Drives Gene Expression and Cancer Progression. Cell 2016;165:1416-27.
[9] Berns A. A tRNA with oncogenic capacity. Cell 2008;133:29-30.
[10] Waldman YY, Tuller T, Sharan R, Ruppin E. TP53 cancerous mutations exhibit selection for translation efficiency. Cancer research 2009;69:8807-13.
[11] Kushner JP, Boll D, Quagliana J, Dickman S. Elevated methionine-tRNA synthetase activity in human colon cancer. Proceedings of the Society for Experimental Biology and Medicine Society for Experimental Biology and Medicine 1976; 153:273-6.
[12] Marshall L, Kenneth NS, White RJ. Elevated tRNA (iMet) synthesis can drive cell proliferation and oncogenic transformation. Cell 2008;133:78-89.
[13] Pavon-Eternod M, Gomes S, Geslain R, Dai Q, Rosner MR, Pan T. tRNA over-expression in breast cancer and functional consequences. Nucleic acids research 2009;37:7268-80.
[14] Zhou Y, Goodenbour JM, Godley LA, Wickrema A, Pan T. High levels of tRNA abundance and alteration of tRNA charging by bortezomib in multiple myeloma. Biochemical and biophysical research communications 2009;385:160-4.
[15] Begley U, Sosa MS, Avivar-Valderas A, Patil A, Endres L, Estrada Y, et al. A human tRNA methyltransferase 9-like protein prevents tumour growth by regulating LIN9 and HIF1-alpha. EMBO molecular medicine 2013; 5:366-83.
[16] Girstmair H, Saffert P, Rode S, Czech A, Holland G, Bannert N, et al. Depletion of cognate charged transfer RNA causes translational frameshifting within the expanded CAG stretch in huntingtin. Cell reports 2013;3:148-59 .
[17] Pavon-Eternod M, David A, Dittmar K, Berglund P, Pan T, Bennink JR, et al. Vaccinia and influenza A viruses select rather than adjust tRNAs to optimize translation. Nucleic acids research 2013;41:1914-21 .
[18] van Weringh A, Ragonnet-Cronin M, Pranckeviciene E, Pavon-Eternod M, Kleiman L, Xia X. HIV-1 modulates the tRNA pool to improve translation efficiency. Molecular biology and evolution 2011;28:1827-34.